table s3: list of genes differentially regulated by 6sialyllactose treatment. gene name description p-value fold change bl
Table S3: List of genes differentially regulated by 6'sialyllactose
treatment.
Gene Name
Description
P-Value
Fold Change
Blon_0028
conserved hypothetical protein
0.0491
1.35
Blon_0029
Ferritin, Dps family protein
0.0002
1.72
Blon_0036
FAD-dependent pyridine nucleotide-disulphide oxidoreductase
0.0365
1.26
Blon_0268
glycoside hydrolase family 2, sugar binding
0.0044
0.71
Blon_0286
lactoylglutathione lyase (LGUL) family protein, diverged
0.0441
1.91
Blon_0291
conserved hypothetical protein
0.0095
1.58
Blon_0307
ATP synthase F1, alpha subunit
0.0147
0.79
Blon_0332
hypothetical protein
0.0270
0.79
Blon_0335
putative transcriptional regulator, MerR family
0.0102
0.78
Blon_0392
cation efflux protein
0.0001
1.84
Blon_0394
glutamine amidotransferase class-I
0.0102
1.61
Blon_0450
hypothetical protein
0.0308
1.31
Blon_0459
glycoside hydrolase, family 20
0.0118
1.65
Blon_0460
binding-protein-dependent transport systems inner membrane component
0.0191
1.38
Blon_0505
hypothetical protein
0.0194
0.79
Blon_0518
hypothetical protein
0.0197
0.74
Blon_0536
hypothetical protein
0.0216
1.26
Blon_0573
ROK family protein
0.0341
0.75
Blon_0615
Resolvase, N-terminal domain protein
0.0260
1.24
Blon_0616
transposase, IS605 OrfB family
0.0446
1.25
Blon_0617
glutamate--cysteine ligase, GCS2
0.0009
1.63
Blon_0619
DNA polymerase, beta domain protein region
0.0014
1.59
Blon_0620
nucleotidyltransferase substrate binding protein, HI0074 family
0.0085
1.49
Blon_0621
Glucan 1,3-beta-glucosidase
0.0462
1.34
Blon_0643
conserved hypothetical protein
0.0308
0.78
Blon_0644
ROK family protein
0.0204
0.73
Blon_0645
N-acylglucosamine-6-phosphate 2-epimerase
0.0344
0.74
Blon_0748
Cystathionine gamma-synthase
0.0484
0.83
Blon_0758
Glutaredoxin-like protein
0.0044
1.45
Blon_0759
ABC transporter related
0.0463
1.37
Blon_0789
periplasmic binding protein/LacI transcriptional regulator
0.0456
0.83
Blon_0790
proteinase inhibitor I4, serpin
0.0330
0.78
Blon_0852
UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate--D-alanyl-D-alanyl
ligase
0.0453
0.82
Blon_0862
ABC transporter related
0.0141
1.34
Blon_0863
ABC-2 type transporter
0.0319
1.23
Blon_0864
ISXoo15 transposase
0.0343
1.24
Blon_0865
putative transcriptional regulator
0.0028
1.52
Blon_0884
binding-protein-dependent transport systems inner membrane component
0.0295
0.77
Blon_0885
binding-protein-dependent transport systems inner membrane component
0.0020
0.72
Blon_0902
initiation factor 3
0.0083
1.59
Blon_0947
helix-turn-helix domain protein
0.0067
1.35
Blon_0948
hypothetical protein
0.0199
1.30
Blon_0991
conserved hypothetical protein
0.0032
1.90
Blon_0992
hypothetical protein
0.0004
1.98
Blon_0993
hypothetical protein
0.0008
1.79
Blon_0994
transcriptional regulator, Fis family
0.0299
1.24
Blon_1037
conserved hypothetical protein
0.0485
1.41
Blon_1205
hypothetical protein Blon_1205
0.0234
0.78
Blon_1315
transposase, IS605 OrfB family
0.0072
1.29
Blon_1495
conserved hypothetical protein
0.0114
1.39
Blon_1496
helix-turn-helix domain protein
0.0095
1.54
Blon_1539
hypothetical protein
0.0392
1.26
Blon_1540
hypothetical protein
0.0293
1.25
Blon_1541
hypothetical protein
0.0186
1.41
Blon_1542
hypothetical protein
0.0218
1.39
Blon_1545
Cpl-7 lysozyme, C-terminal domain protein
0.0364
1.28
Blon_1664
GCN5-related N-acetyltransferase
0.0463
1.41
Blon_1687
TfoX, C-terminal domain protein
0.0006
1.93
Blon_1688
transcription activator, effector binding
0.0000
2.35
Blon_1689
GTP-binding protein YchF
0.0200
1.41
Blon_1691
proline iminopeptidase
0.0064
1.31
Blon_1692
integral membrane sensor signal transduction histidine kinase
0.0004
1.83
Blon_1693
two component transcriptional regulator, LuxR family
0.0007
1.72
Blon_1697
Phosphomethylpyrimidine kinase type-1
0.0020
1.74
Blon_1698
protein of unknown function UPF0102
0.0006
1.82
Blon_1700
SMF family protein
0.0422
1.26
Blon_1713
narrowly conserved hypothetical protein
0.0063
0.74
Blon_1714
pyruvate formate-lyase activating enzyme
0.0252
0.80
Blon_1761
1,4-alpha-glucan branching enzyme
0.0241
0.77
Blon_1902
conserved hypothetical protein
0.0035
0.73
Blon_1910
conserved hypothetical protein
0.0103
1.34
Blon_1950
hypothetical protein
0.0313
1.20
Blon_1951
UMUC domain protein DNA-repair protein
0.0332
1.22
Blon_2061
extracellular solute-binding protein, family 1
0.0254
1.29
Blon_2064
transcriptional regulator, DeoR family
0.0334
1.24
Blon_2081
conserved hypothetical protein
0.0199
1.29
Blon_2082
lipopolysaccharide biosynthesis
0.0112
1.50
Blon_2173
aminoglycoside phosphotransferase
0.0236
0.79
Blon_2174
conserved hypothetical protein
0.0060
0.64
Blon_2175
binding-protein-dependent transport systems inner membrane component
0.0033
0.64
Blon_2176
binding-protein-dependent transport systems inner membrane component
0.0013
0.62
Blon_2186
narrowly conserved hypothetical protein
0.0345
1.27
Blon_2191
ribose 5-phosphate isomerase
0.0158
1.25
Blon_2335
conserved hypothetical protein
0.0427
0.83
Blon_2341
protein of unknown function DUF624
0.0113
0.75
Blon_2342
binding-protein-dependent transport systems inner membrane component
0.0086
0.76
Blon_2348
Exo-alpha-sialidase
0.0355
0.78
Blon_2349
dihydrodipicolinate synthetase
0.0487
0.83
Blon_2370
glycerophosphoryl diester phosphodiesterase
0.0001
1.83
Blon_2371
Glutamate--tRNA ligase
0.0001
1.75
Blon_2372
ATPase AAA-2 domain protein
0.0000
1.88
Blon_2379
binding-protein-dependent transport systems inner membrane component
0.0221
0.70
Blon_2380
extracellular solute-binding protein, family 1
0.0115
0.73
dnaK
chaperone protein DnaK
0.0005
1.58
groEL
chaperonin GroEL
0.0147
1.37
recA
recA protein
0.0135
1.26
rpmI
ribosomal protein L35
0.0055
1.47
thiG
thiazole biosynthesis family protein
0.04100675
0.80
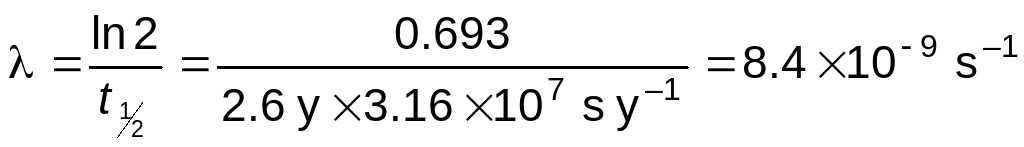 TAP 5162 RADIOACTIVE DECAY WITH EXPONENTIALS 1 THE HALFLIFE
TAP 5162 RADIOACTIVE DECAY WITH EXPONENTIALS 1 THE HALFLIFE MINISTARSTVO ZDRAVLJA 1109 NA TEMELJU ČLANKA 12 STAVKA 3
MINISTARSTVO ZDRAVLJA 1109 NA TEMELJU ČLANKA 12 STAVKA 3 LA CALIDAD EN LOS CENTROS DE FORMACIÓN CRITERIOS E
LA CALIDAD EN LOS CENTROS DE FORMACIÓN CRITERIOS E MARCHES PUBLICS DECLARATION DE SOUSTRAITANCE DC4 LE FORMULAIRE DC4
MARCHES PUBLICS DECLARATION DE SOUSTRAITANCE DC4 LE FORMULAIRE DC4 DEPARTAMENT DE MATEMÀTIQUES IES – JMª QUADRADO CIUTADELLA
DEPARTAMENT DE MATEMÀTIQUES IES – JMª QUADRADO CIUTADELLA VARSEL I HENHOLD TIL NS 3430 – PKT 175
VARSEL I HENHOLD TIL NS 3430 – PKT 175 COMUNICADO DE PRENSA LOS SINDICATOS DENUNCIAN UN NUEVO
COMUNICADO DE PRENSA LOS SINDICATOS DENUNCIAN UN NUEVO 7 APPLICATION FORM FOR THE GRADUATE SCHOLARSHP PROGRAM ME
7 APPLICATION FORM FOR THE GRADUATE SCHOLARSHP PROGRAM ME